1 A crash course in R and RStudio
In this section, we’re going to very briefly go over the basics of R. This is meant to get everyone on the same page. If you’ve done the pre-work we described on the tutorial github repo, this will hopefully be pretty familiar!
1.1 R vs RStudio
In this workshop we are going to be learning the basics of coding for text analysis in R, but we will be using the RStudio interface/IDE! Why am I using R for this workshop? And why do we need this extra layer of program to deal with it?
1.1.1 What is R?
R is a programming language that was built for statistical and numerical analysis. It is not unique in these spaces–most of you are probably familiar with a program like SAS, SPSS, Unscrambler, XLSTAT, JMP, etc. Unlike these, R is free and open-source. This has two main consequences:
Ris constantly being extended to do new, useful things because there is a vibrant community of analysts developing tools for it, and the barrier to entry is very low.
Rdoesn’t have a fixed set of tasks that it can accomplish, and, in fact, I generally haven’t found a data-analysis task I needed to do that I couldn’t inR.
Because it’s a programming language, R isn’t point-and-click–today we’re going to be typing commands into the console, hitting, enter, making errors, and repeating. But this is a good thing! The power and flexibility of R (and its ability to do most of the things we want) come from the fact that it is a programming language. While learning to use R can seem intimidating, the effort to do so will give you a much more powerful suite of tools than the more limited point-and-click alternatives. R is built for research programming (data analysis), rather than for production programming. The only other alternative that is as widely supported in the research community is Python, but–honesty time here–I have never learned Python very well, and so we are learning R. And, in addition, Python doesn’t have as good an Interactive Development Environment (IDE, explained further below) as RStudio!
If you open your R.exe/R application, you’ll see something like this:
R graphical consoleYou can also work with R from a shell interface, but I will not be discussing this approach in this workshop.
1.1.2 Great, why are we using RStudio then?
RStudio is an “Interactive Development Environment” (IDE) for working with R. Without going into a lot of detail, that means that R lives on its own on your computer in a separate directory, and RStudio provides a bunch of better functionality for things like writing multiple files at once, making editing easier, autofilling code, and displaying plots. You can learn more about RStudio here.
With that out of the way, I am going to be sloppy in terminology and say/type “R” a lot of the times I mean “RStudio”. I will be very clear if the distinction actually matters. RStudio is going to make your life way easier, and when you try to learn Python you are going to be sad :(
1.2 The parts of RStudio
The default layout of RStudio looks something like this (font sizes may vary):
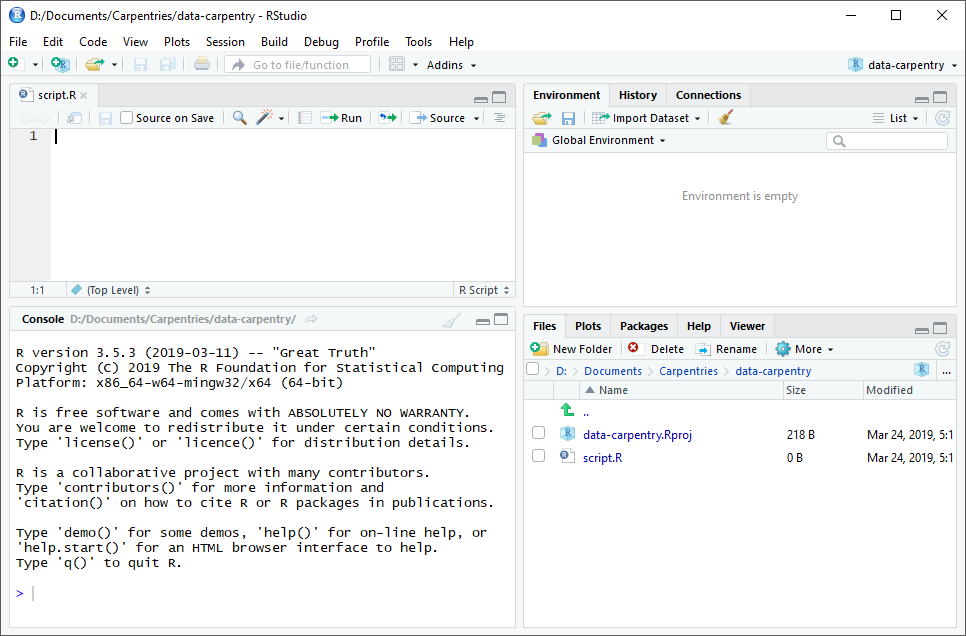
RStudio always has 4 “panes” with various functions, which are in tabs (just like a web browser). The key ones for right now to pay attention are:
- The
Consoletab is the portal to interact directly withR. The>“prompt” is where you can type and execute commands (by hitting return). You can try this out right now by using it like a calculator - try1 + 1if you like! - The
Filestab shows the files in your working directory: like in the Windows Explorer or macOS Finder, files are displayed within folders. You can click on files to open them. - The
Helptab shows documentation forRfunctions and packages–it is useful for learning how to use specific functions. - The
Plotstab shows graphical output, and this is where the data visualizations we’ll learn to make will (generally) appear. - The
Environmenttab shows the objects that exist in memory in your currentRsession. Without going into details, this is “what you’ve done” so far: data tables and variables you’ve created, etc. - Finally, the
Scriptspane shows individual tabs for each script and other RStudio file. Scripts (and other, more exotic file types like RMarkdown/.Rmd files) are documents that contain multipleRcommands, like you’d type into theConsole. However, unlike commands in theConsole, these commands don’t disappear as soon as they’re run, and we can string them together to make workflows or even programs. This is where the real power ofRwill come from.
You can change the layout of your Panes (and many other options) by going to the RStudio menu: Tools > Global Options and select Pane Layout.
You’ll notice that my layout for RStudio looks quite different from the default, but you can always orient yourself by seeing what tab or pane I am in–these are always the same. I prefer giving myself more space for writing R scripts and markdown files, so I have given that specific Pane more space while minimizing the History pane.
While we’re in Global Options, please make the following selections:
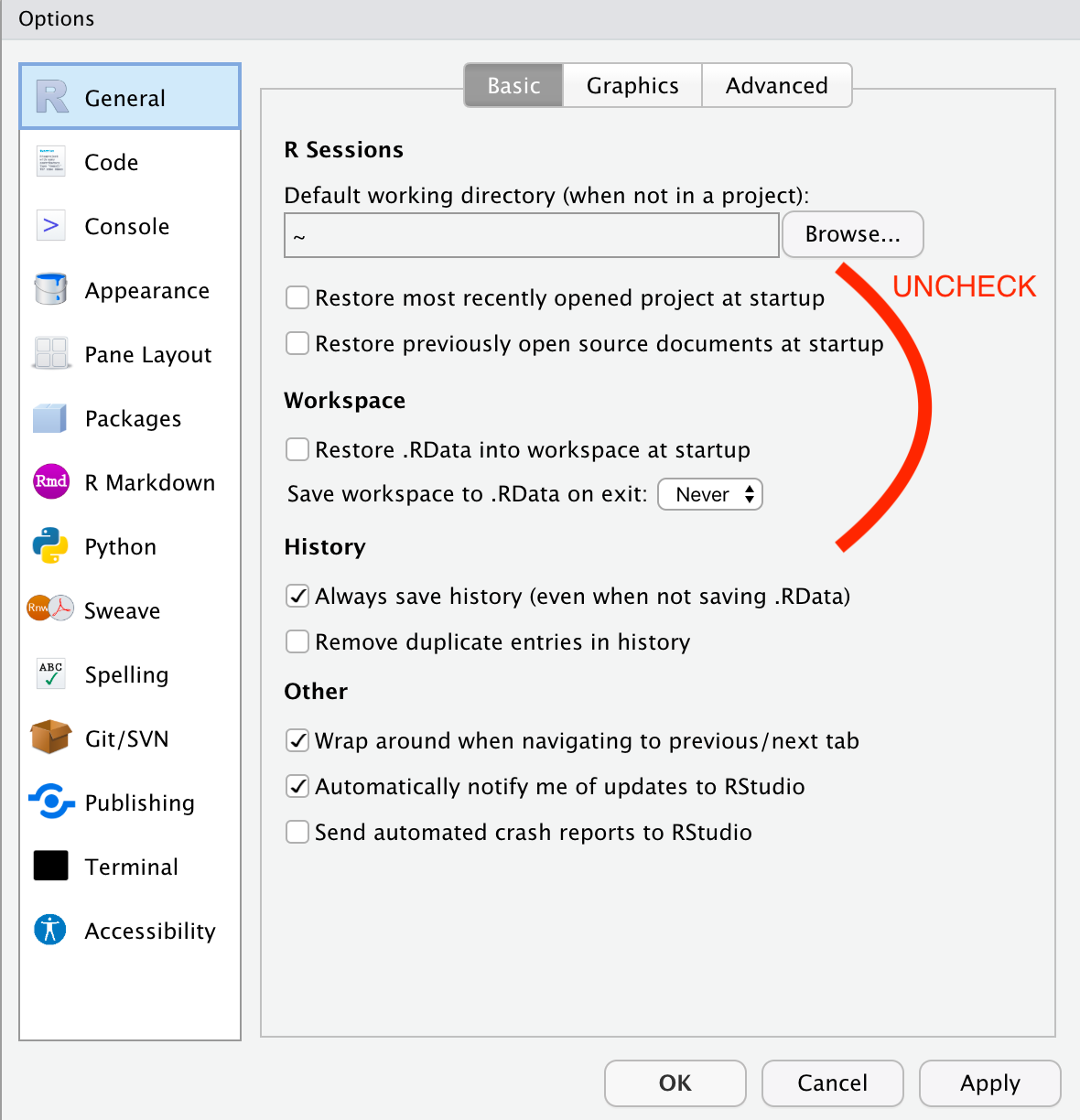
- Under
General, uncheck all the boxes to do with restoring projects and workspaces. We want to make sure our code runs the same time every time (i.e., that our methods are reproducible), and letting RStudio load these will make this impossible:
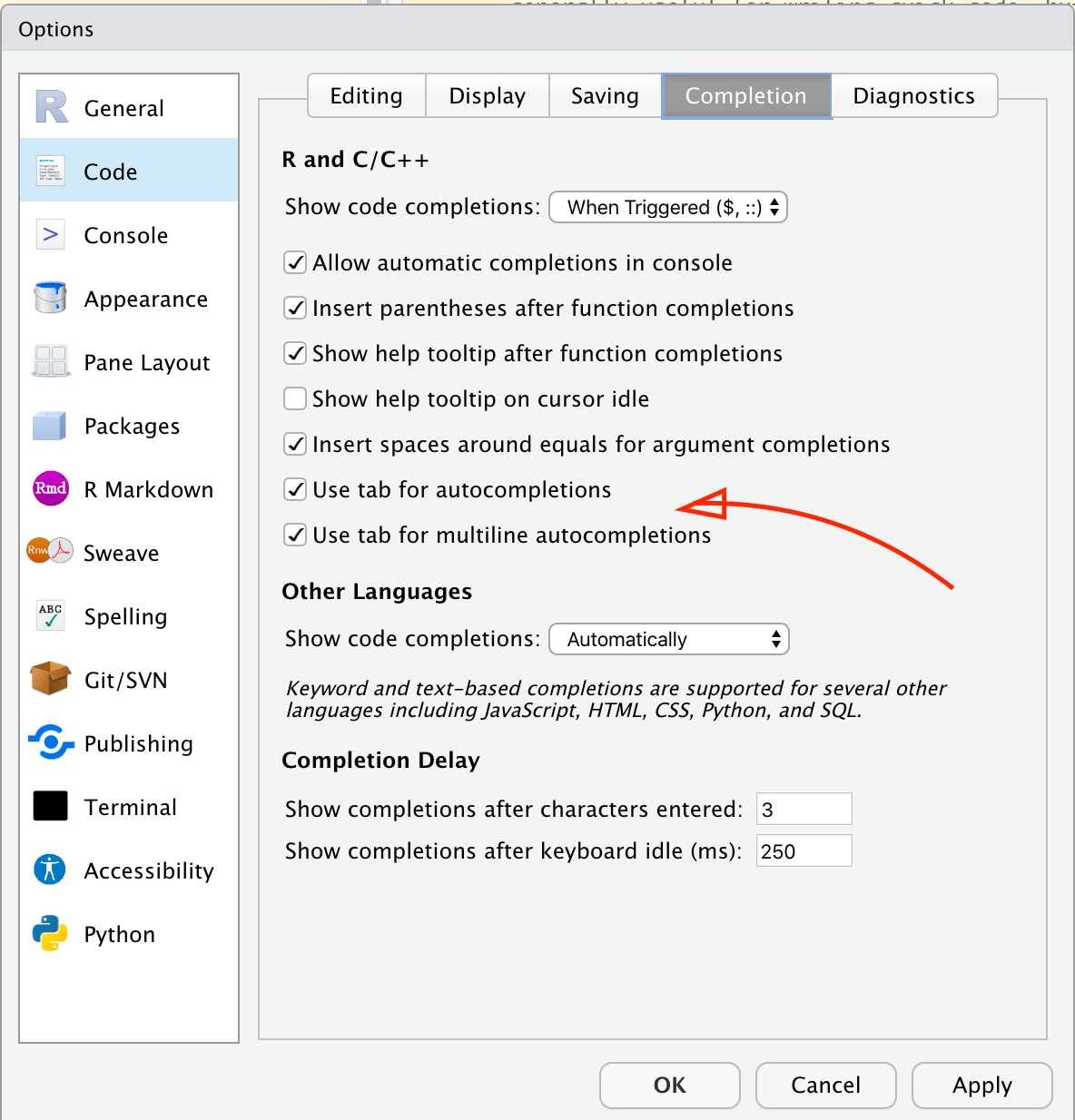
- Make your life easier by setting up autocompletion for your code. Under the
Code > Completionoptions, select the checkboxes to allow usingtabfor autocompletions, and also allowing multiline autocompletions. This means that RStudio will suggest functions and data for you if you hittab, which will make you have to do way less typing:
1.2.1 The “working directory” and why you should care
Before we move on to using R for real, we have one key general computing concept to tackle: the “working directory”. The working directory is the folder on your computer in which R will look for files and save files.
When you need to tell R to read in data from a file or output a file, you will have to do so in relation to your working directory. Therefore, it is important that you know how to find your working directory and change it.
The easiest (but not best) way to do this is to use the Files pane. If you hit the “gear” icon in the Files pane menu, you’ll see two commands to do with the working directory. You can Go To Working Directory to show you whatever R currently has set as the working directory. You can then navigate to any directory you want on your hard drive, and use the Set As Working Directory command to make that the working directory.
A better way to do this is to use the R commands getwd() and setwd().
## [1] "C:/Users/lhamilton/Documents/sensometrics-r-tutorial-2024"And we can manually change the working directory by using
Notice that I am not running the second command, because it would cause an error!
When we use R to navigate directories, I recommend always using the forward slash: /, even though on Windows systems the typical slash is the backslash: \. R will properly interpret the / for you in the context of your operating system, and this is more consistent with most modern code environments.
1.3 Extending R with packages
One of the key advantages of R is that its open-source nature means that you can extend it to do all sorts of things. For example, for much of this workshop we are going to use the tidyverse packages and the ca package. There are various ways to install new packages, but the easiest way is to use the Packages tab. This will show you all the packages you currently have installed as an alphabetical list.
We install new packages using the install.packages() command. As part of the pre-work for the tutorial, you will have run the following commmands:
# Install the tidyverse packages
install.packages("tidyverse")
# Install the ca package
install.packages("ca")While these commands will download packages and install them in a directory on your hard drive where R can find them, they will not make them immediately usable in an R programming session.
1.3.1 Loading packages
To actually use a package, you need to load it using the library(<name of package>) command. So, for example, to load the tidyverse package we will use the command
## ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
## ✔ dplyr 1.1.4 ✔ readr 2.1.4
## ✔ forcats 1.0.0 ✔ stringr 1.5.1
## ✔ ggplot2 3.4.2 ✔ tibble 3.2.1
## ✔ lubridate 1.9.2 ✔ tidyr 1.3.1
## ✔ purrr 1.0.1
## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
## ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsYou need to use multiple library() commands to load multiple packages, e.g.,
If you want to know what packages you have loaded, you can run the sessionInfo() function, which will tell you a bunch of stuff, including the “attached” packages:
## R version 4.3.1 (2023-06-16 ucrt)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 19045)
##
## Matrix products: default
##
##
## locale:
## [1] LC_COLLATE=English_United States.utf8
## [2] LC_CTYPE=English_United States.utf8
## [3] LC_MONETARY=English_United States.utf8
## [4] LC_NUMERIC=C
## [5] LC_TIME=English_United States.utf8
##
## time zone: America/New_York
## tzcode source: internal
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ca_0.71.1 lubridate_1.9.2 forcats_1.0.0 stringr_1.5.1
## [5] dplyr_1.1.4 purrr_1.0.1 readr_2.1.4 tidyr_1.3.1
## [9] tibble_3.2.1 ggplot2_3.4.2 tidyverse_2.0.0
##
## loaded via a namespace (and not attached):
## [1] gtable_0.3.3 jsonlite_1.8.8 compiler_4.3.1 tidyselect_1.2.1
## [5] jquerylib_0.1.4 scales_1.3.0 yaml_2.3.7 fastmap_1.1.1
## [9] R6_2.5.1 generics_0.1.3 knitr_1.43 bookdown_0.34
## [13] munsell_0.5.1 bslib_0.7.0 pillar_1.9.0 tzdb_0.4.0
## [17] rlang_1.1.1 utf8_1.2.4 stringi_1.7.12 cachem_1.0.8
## [21] xfun_0.39 sass_0.4.9 timechange_0.2.0 cli_3.6.1
## [25] withr_3.0.0 magrittr_2.0.3 digest_0.6.33 grid_4.3.1
## [29] rstudioapi_0.15.0 hms_1.1.3 lifecycle_1.0.4 vctrs_0.6.5
## [33] evaluate_0.23 glue_1.6.2 fansi_1.0.6 colorspace_2.1-0
## [37] rmarkdown_2.23 tools_4.3.1 pkgconfig_2.0.3 htmltools_0.5.8.1Finally, you can also load (and unload) packages using the Packages tab, by clicking the checkbox next to the name of the package you want to load (or unload).
Typically, we think it’s a good idea to put all of your library() function calls at the top of your script, so that you don’t try to call a function from a package that has been loaded “later” (further down) in your script and cause an error.
1.4 Making R do stuff
We can make R act as a basic calculator:
## [1] 5## [1] 0.03225806Mostly, we will be using R for writing complex, functional programs, in which we assign variables and use those variables in functions:
## [1] "You've got something else there!"1.5 Livecoding along
We’ve now covered the Console tab and the Scripts pane. These are both areas in which you can write and execute code, but they work a little differently. The Console is the place to run code that is short and easy to type, or that you’re experimenting with. It will allow you to write a single line of code, and after you hit return, R will execute the command. This is great for “interactive programming”, but it isn’t so great for building up a complex workflow, or for following along with this workshop!
This is why I have recommended that you create a new script to follow along with this workshop. You get a new script by going to File > New File > R Script. You can write multiple lines of code and then execute each one in any order (although keeping a logical sequence from top to bottom will help you keep track of what you’re doing). In an R script, everything is expected to be valid R code.
You can't write this in an R script because it is plain text. This will
cause an error.
# If you want to write text or notes to yourself, use the "#" symbol at the start of
# every line to "comment" out that line. You can also put "#" in the middle of
# a line in order to add a comment - everything after will be ignored.
1 + 1 # this is valid R syntax
print("hello world") # this is also valid R syntaxTo run code from your R script, put your cursor on the line you want to run and either hit the run button with the green arrow at the top left or (my preferred method) type cmd + return (on Mac) or ctrl + return (on PC).